Featurization Model Selection and Tuning
Overview
The data contains features extracted from the silhouette of vehicles in different angles. The purpose is to classify a given silhouette as one of three types of vehicle, using a set of features extracted from the silhouette. The vehicle may be viewed from one of many different angles.
Solution
Importing Necessary libraries
1# NumPy: For mathematical funcations, array, matrices operations
2import numpy as np
3
4# Graph: Plotting graphs and other visula tools
5import pandas as pd
6import seaborn as sns
7
8sns.set(color_codes=True)
9
10#To enable inline plotting graphs
11import matplotlib.pyplot as plt
12%matplotlib inline
13
14# Disable warning
15import warnings
16warnings.filterwarnings("ignore")
Load Data
1# Load data set
2# Import CSV data using pandas data frame
3df_original = pd.read_csv('concrete.csv')
4
5# Print total columns
6print("Total Colums in dataframe: ", len(df_original.columns))
7
8# Prepare columns names
9df_original_columns = []
10for column in df_original.columns:
11 df_original_columns.append(column)
12
13
14
15print("Columns list {}".format(df_original_columns))
16print("***********************************************************************************************************************")
17
18# Prepare mapping of column names for quick access
19df_original_columns_map = {}
20map_index: int = 0
21for column in df_original_columns:
22 df_original_columns_map[map_index] = column
23 map_index = map_index + 1
24
25print("Columns Map {}".format(df_original_columns_map))
Total Colums in dataframe: 9
Columns list ['cement', 'slag', 'ash', 'water', 'superplastic', 'coarseagg', 'fineagg', 'age', 'strength']
***********************************************************************************************************************
Columns Map {0: 'cement', 1: 'slag', 2: 'ash', 3: 'water', 4: 'superplastic', 5: 'coarseagg', 6: 'fineagg', 7: 'age', 8: 'strength'}
Data Pre-Processing
Data Shape
1df_original.shape
(1030, 9)
Data Info
1df_original.info()
2# All row items are numeric in nature, hence no label encoding is required
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 1030 entries, 0 to 1029
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 cement 1030 non-null float64
1 slag 1030 non-null float64
2 ash 1030 non-null float64
3 water 1030 non-null float64
4 superplastic 1030 non-null float64
5 coarseagg 1030 non-null float64
6 fineagg 1030 non-null float64
7 age 1030 non-null int64
8 strength 1030 non-null float64
dtypes: float64(8), int64(1)
memory usage: 72.5 KB
Data
1df_original.head(16)
| cement | slag | ash | water | superplastic | coarseagg | fineagg | age | strength | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 141.3 | 212.0 | 0.0 | 203.5 | 0.0 | 971.8 | 748.5 | 28 | 29.89 |
| 1 | 168.9 | 42.2 | 124.3 | 158.3 | 10.8 | 1080.8 | 796.2 | 14 | 23.51 |
| 2 | 250.0 | 0.0 | 95.7 | 187.4 | 5.5 | 956.9 | 861.2 | 28 | 29.22 |
| 3 | 266.0 | 114.0 | 0.0 | 228.0 | 0.0 | 932.0 | 670.0 | 28 | 45.85 |
| 4 | 154.8 | 183.4 | 0.0 | 193.3 | 9.1 | 1047.4 | 696.7 | 28 | 18.29 |
| 5 | 255.0 | 0.0 | 0.0 | 192.0 | 0.0 | 889.8 | 945.0 | 90 | 21.86 |
| 6 | 166.8 | 250.2 | 0.0 | 203.5 | 0.0 | 975.6 | 692.6 | 7 | 15.75 |
| 7 | 251.4 | 0.0 | 118.3 | 188.5 | 6.4 | 1028.4 | 757.7 | 56 | 36.64 |
| 8 | 296.0 | 0.0 | 0.0 | 192.0 | 0.0 | 1085.0 | 765.0 | 28 | 21.65 |
| 9 | 155.0 | 184.0 | 143.0 | 194.0 | 9.0 | 880.0 | 699.0 | 28 | 28.99 |
| 10 | 151.8 | 178.1 | 138.7 | 167.5 | 18.3 | 944.0 | 694.6 | 28 | 36.35 |
| 11 | 173.0 | 116.0 | 0.0 | 192.0 | 0.0 | 946.8 | 856.8 | 3 | 6.94 |
| 12 | 385.0 | 0.0 | 0.0 | 186.0 | 0.0 | 966.0 | 763.0 | 14 | 27.92 |
| 13 | 237.5 | 237.5 | 0.0 | 228.0 | 0.0 | 932.0 | 594.0 | 7 | 26.26 |
| 14 | 167.0 | 187.0 | 195.0 | 185.0 | 7.0 | 898.0 | 636.0 | 28 | 23.89 |
| 15 | 213.8 | 98.1 | 24.5 | 181.7 | 6.7 | 1066.0 | 785.5 | 100 | 49.97 |
Data Description
1df_original.describe().T
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| cement | 1030.0 | 281.167864 | 104.506364 | 102.00 | 192.375 | 272.900 | 350.000 | 540.0 |
| slag | 1030.0 | 73.895825 | 86.279342 | 0.00 | 0.000 | 22.000 | 142.950 | 359.4 |
| ash | 1030.0 | 54.188350 | 63.997004 | 0.00 | 0.000 | 0.000 | 118.300 | 200.1 |
| water | 1030.0 | 181.567282 | 21.354219 | 121.80 | 164.900 | 185.000 | 192.000 | 247.0 |
| superplastic | 1030.0 | 6.204660 | 5.973841 | 0.00 | 0.000 | 6.400 | 10.200 | 32.2 |
| coarseagg | 1030.0 | 972.918932 | 77.753954 | 801.00 | 932.000 | 968.000 | 1029.400 | 1145.0 |
| fineagg | 1030.0 | 773.580485 | 80.175980 | 594.00 | 730.950 | 779.500 | 824.000 | 992.6 |
| age | 1030.0 | 45.662136 | 63.169912 | 1.00 | 7.000 | 28.000 | 56.000 | 365.0 |
| strength | 1030.0 | 35.817961 | 16.705742 | 2.33 | 23.710 | 34.445 | 46.135 | 82.6 |
Checking for Missing value, duplicate data, incorrect data and perform data cleansing
Empty NA Values
1df_original.isna().sum()
2# No null values are present in data
cement 0
slag 0
ash 0
water 0
superplastic 0
coarseagg 0
fineagg 0
age 0
strength 0
dtype: int64
Duplicates
1df_duplicates = df_original.duplicated()
2
3print('Number of duplicate rows = {}'.format(df_duplicates.sum()))
4
5# 25 duplicates
6
7# We are not modyfying original dataframe instead our all operations
8# will be on `df_main`
9df_main = df_original.drop_duplicates()
10
11df_main.shape
Number of duplicate rows = 25
(1005, 9)
Pearson Correlation
1df_main.corr(method ='pearson')
2# Eg. Present of cement, superplastic and more value of age result more strength of the mixture
| cement | slag | ash | water | superplastic | coarseagg | fineagg | age | strength | |
|---|---|---|---|---|---|---|---|---|---|
| cement | 1.000000 | -0.303324 | -0.385610 | -0.056625 | 0.060906 | -0.086205 | -0.245375 | 0.086348 | 0.488283 |
| slag | -0.303324 | 1.000000 | -0.312352 | 0.130262 | 0.019800 | -0.277559 | -0.289685 | -0.042759 | 0.103374 |
| ash | -0.385610 | -0.312352 | 1.000000 | -0.283314 | 0.414213 | -0.026468 | 0.090262 | -0.158940 | -0.080648 |
| water | -0.056625 | 0.130262 | -0.283314 | 1.000000 | -0.646946 | -0.212480 | -0.444915 | 0.279284 | -0.269624 |
| superplastic | 0.060906 | 0.019800 | 0.414213 | -0.646946 | 1.000000 | -0.241721 | 0.207993 | -0.194076 | 0.344209 |
| coarseagg | -0.086205 | -0.277559 | -0.026468 | -0.212480 | -0.241721 | 1.000000 | -0.162187 | -0.005264 | -0.144717 |
| fineagg | -0.245375 | -0.289685 | 0.090262 | -0.444915 | 0.207993 | -0.162187 | 1.000000 | -0.156572 | -0.186448 |
| age | 0.086348 | -0.042759 | -0.158940 | 0.279284 | -0.194076 | -0.005264 | -0.156572 | 1.000000 | 0.337367 |
| strength | 0.488283 | 0.103374 | -0.080648 | -0.269624 | 0.344209 | -0.144717 | -0.186448 | 0.337367 | 1.000000 |
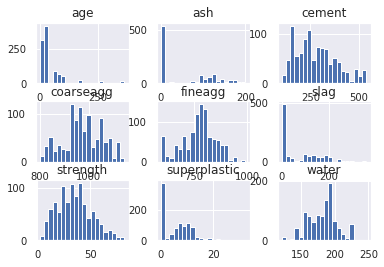
Body of Distribution
1df_main.hist(bins=20, xlabelsize=10, ylabelsize=10)
2# We can see that data is not distributed properly and there are tails present on either side of gaussian curve
array([[<matplotlib.axes._subplots.AxesSubplot object at 0x7faef7133a50>,
<matplotlib.axes._subplots.AxesSubplot object at 0x7faef68cf990>,
<matplotlib.axes._subplots.AxesSubplot object at 0x7faef688dd10>],
[<matplotlib.axes._subplots.AxesSubplot object at 0x7faef68419d0>,
<matplotlib.axes._subplots.AxesSubplot object at 0x7faef67fed50>,
<matplotlib.axes._subplots.AxesSubplot object at 0x7faef67b2a10>],
[<matplotlib.axes._subplots.AxesSubplot object at 0x7faef6772e90>,
<matplotlib.axes._subplots.AxesSubplot object at 0x7faef67a6a50>,
<matplotlib.axes._subplots.AxesSubplot object at 0x7faef67315d0>]],
dtype=object)
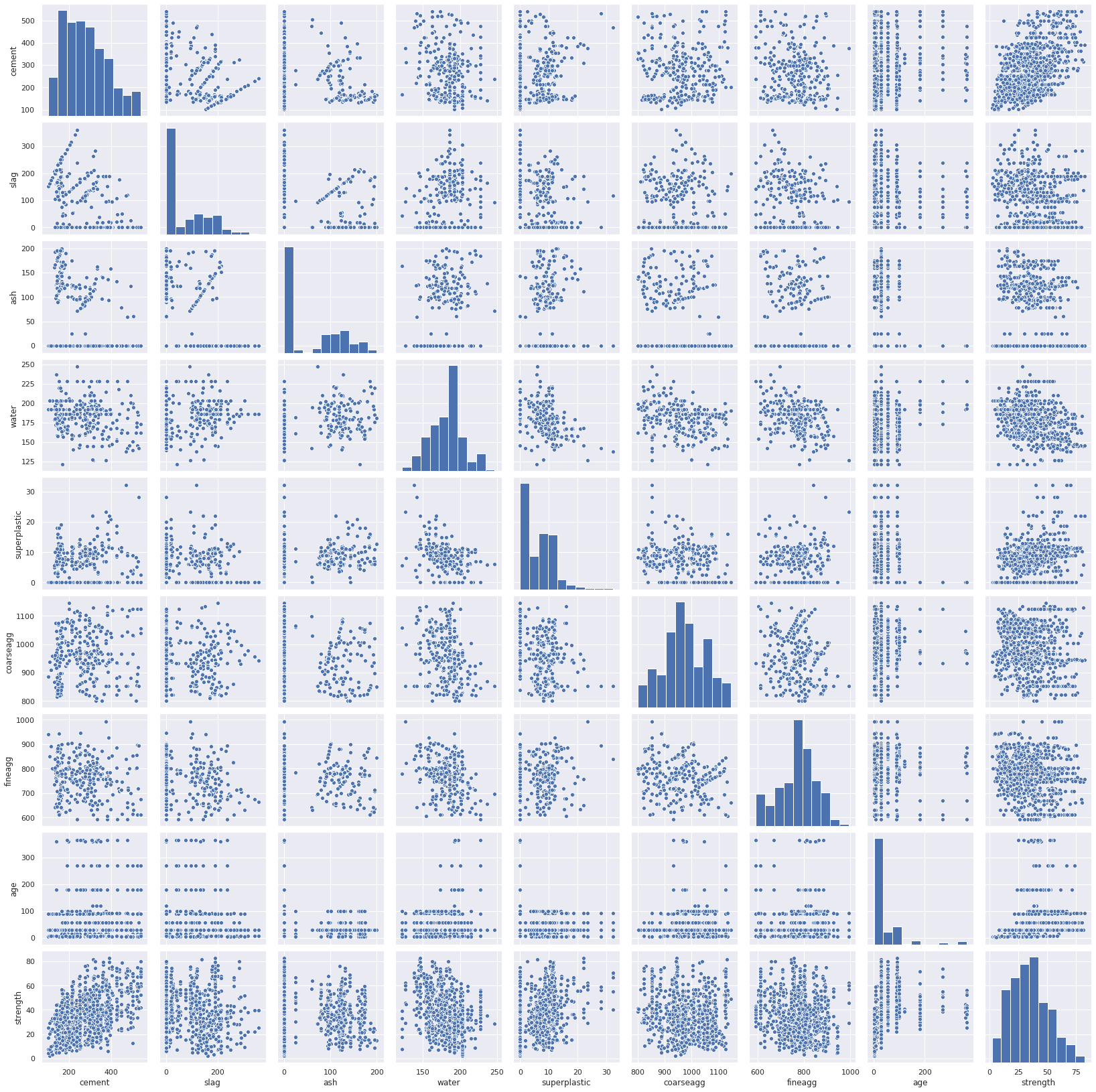
Pairplot
1sns.pairplot(df_main)
2# We do not see any positive curve or negative curve but there is a cloud
3# which indicates the presence of cement add strength though not direct relation
<seaborn.axisgrid.PairGrid at 0x7faef6441c90>
Evaluation of different models
1from sklearn.model_selection import cross_val_score
2from sklearn.model_selection import train_test_split
3from sklearn.pipeline import Pipeline
4from sklearn.model_selection import KFold, GridSearchCV, RandomizedSearchCV
5from sklearn.preprocessing import MinMaxScaler
6from sklearn.linear_model import LinearRegression, Ridge,Lasso
7from sklearn.neighbors import KNeighborsRegressor
8from sklearn.ensemble import AdaBoostRegressor, ExtraTreesRegressor, RandomForestRegressor, GradientBoostingRegressor
9from sklearn.tree import DecisionTreeRegressor
10
11
12class ModelEvaluator:
13
14 def __init__(self, dataFrame):
15 self.dataFrame = dataFrame
16 self.modelResullt = {}
17 self.algorithms = {
18 "linearRegression" : LinearRegression(),
19 "lasso" : Lasso(),
20 "ridge" : Ridge(),
21 "adaBoostRegressor" : AdaBoostRegressor(),
22 "extraTreeRegressor" : ExtraTreesRegressor(),
23 "randomForestRegressor" : RandomForestRegressor(),
24 "gradientBoostRegressor" : GradientBoostingRegressor(),
25 "decisionTreeRegressor" : DecisionTreeRegressor(),
26 "knnRegressor" : KNeighborsRegressor()
27 }
28
29 def printDf_X(self):
30 print(self.df_X)
31
32 def printDf_Y(self):
33 print(self.df_Y)
34
35 def splitData(self):
36 print("Splitting data into train ,test and validation")
37 test_size = 0.2
38 seed = 29
39 self.df_X = self.dataFrame.copy().drop(['strength'], axis = 1)
40 self.df_Y = self.dataFrame['strength']
41
42 self.X_train, self.X_test, self.y_train, self.y_test = \
43 train_test_split(self.df_X, self.df_Y, test_size=test_size, random_state=seed)
44
45 self.X_train, self.X_val, self.y_train, self.y_val = \
46 train_test_split(self.df_X, self.df_Y, test_size=test_size, random_state=seed)
47
48
49 def runSimplePipelines(self):
50 print("Running simple pipelines")
51
52 self.decentModels = {}
53
54 for item in self.algorithms:
55 # Init pipe
56 itemPipe = Pipeline([("scaler", MinMaxScaler()), (item, self.algorithms[item])])
57
58 #fit data
59 itemPipe.fit(self.X_train, self.y_train)
60
61 print ("********** {} ************".format(item))
62
63 print("")
64 score = itemPipe.score(self.X_test, self.y_test)
65 print("Test score is {:.2f}". format(score))
66 print("")
67
68 cross_val = cross_val_score(self.algorithms[item], self.X_train, self.y_train)
69 cross_val = cross_val.ravel()
70 print ("cross validatioion score")
71 print ("cv-mean :",cross_val.mean())
72 print ("cv-std :",cross_val.std())
73 print ("cv-max :",cross_val.max())
74 print ("cv-min :",cross_val.min())
75 print("")
76
77 # We aree considering 0.85 s the minim score we need for our prediction
78 if score >= 0.85:
79 self.decentModels[item] = score
80
81 def printDecentModels(self):
82 print("Top performing models are: ")
83 for topModel in self.decentModels:
84 print("{} score {}".format(topModel, self.decentModels[topModel]))
85
86 def deecentModels(self):
87 return selff.decentModels
88
89 def runGridSearchCV_HyperParameterTuningPipelines(self, param1, param2):
90 print("Running gridsearch cv hyper parameter pipelines")
91 for topModel in self.decentModels:
92
93 # Init pipe
94 regressor = self.algorithms[topModel]
95
96 itemPipe = Pipeline([("scaler", MinMaxScaler()), (topModel, regressor)])
97
98 if topModel == 'extraTreeRegressor' or topModel == 'randomForestRegressor':
99
100 gsCv = GridSearchCV(estimator = regressor, param_grid = param1,
101 cv = 3, n_jobs = 1, verbose = 0, return_train_score=True)
102 #fit data
103 gsCv.fit(self.X_train, self.y_train)
104
105 print ("********** {} ************".format(topModel))
106
107 print("")
108
109 print("Best parameter {} ".format(gsCv.best_params_))
110
111 best_grid = gsCv.best_estimator_
112
113 score = best_grid.score(self.X_val, self.y_val)
114
115 print("Best score is {} ".format(score))
116 elif topModel == 'decisionTreeRegressor':
117 gsCv = GridSearchCV(estimator = regressor, param_grid = param2,
118 cv = 3, n_jobs = 1, verbose = 0, return_train_score=True)
119 #fit data
120 gsCv.fit(self.X_train, self.y_train)
121
122 print ("********** {} ************".format(topModel))
123
124 print("")
125
126 print("Best parameter {} ".format(gsCv.best_params_))
127
128 best_grid = gsCv.best_estimator_
129
130 score = best_grid.score(self.X_val, self.y_val)
131
132 print("Best score is {} ".format(score))
133
134
135 def runRandomSearchCV_HyperParameterTuningPipelines(self, param1, param2):
136 print("Running randomsearch cv hyper parameter pipelines")
137 for topModel in self.decentModels:
138 print("")
139 print ("********** {} ************".format(topModel))
140
141 # Init pipe
142 regressor = self.algorithms[topModel]
143
144 itemPipe = Pipeline([("scaler", MinMaxScaler()), (topModel, regressor)])
145
146 if topModel == 'extraTreeRegressor' or topModel == 'randomForestRegressor':
147
148 randCv = RandomizedSearchCV(estimator = regressor, param_distributions = param1,
149 cv = 3, n_jobs = 1, verbose = 0, return_train_score=True)
150 #fit data
151 randCv.fit(self.X_train, self.y_train)
152
153
154 print("Best parameter {} ".format(randCv.best_params_))
155
156 best_random = randCv.best_estimator_
157
158 score = best_random.score(self.X_val, self.y_val)
159
160 print("Best score is {} ".format(score))
161
162 elif topModel == 'gradientBoostRegressor':
163 randCv = RandomizedSearchCV(estimator = regressor, param_distributions = param2,
164 cv = 3, n_jobs = 1, verbose = 0, return_train_score=True)
165 #fit data
166 randCv.fit(self.X_train, self.y_train)
167
168 print("Best parameter {} ".format(randCv.best_params_))
169
170 best_random = randCv.best_estimator_
171
172 score = best_random.score(self.X_val, self.y_val)
173 print("Best score is {} ".format(score))
174
175
1# Instantiation of Class
2modelEvaluator = ModelEvaluator(df_main)
3
4# Splitting data in train, test, validation
5modelEvaluator.splitData()
6
7# Print Data X
8# modelEvaluator.printDf_X()
9
10# Print Data Y
11
12#modelEvaluator.printDf_Y()
Splitting data into train ,test and validation
Run Pipelines
1# run simple pipelines
2
3modelEvaluator.runSimplePipelines()
Running simple pipelines
********** linearRegression ************
Test score is 0.61
cross validatioion score
cv-mean : 0.5814937255975108
cv-std : 0.06146956560114513
cv-max : 0.6549569865710905
cv-min : 0.4825048517025202
********** lasso ************
Test score is 0.18
cross validatioion score
cv-mean : 0.5813839462037981
cv-std : 0.06102069720851994
cv-max : 0.6550985629358492
cv-min : 0.48262897974523195
********** ridge ************
Test score is 0.61
cross validatioion score
cv-mean : 0.5814938391690567
cv-std : 0.06146940364558386
cv-max : 0.654957330984628
cv-min : 0.48250514279718304
********** adaBoostRegressor ************
Test score is 0.79
cross validatioion score
cv-mean : 0.7800716089672267
cv-std : 0.019490295991580992
cv-max : 0.7994746066655304
cv-min : 0.7477560683539344
********** extraTreeRegressor ************
Test score is 0.92
cross validatioion score
cv-mean : 0.895111981820915
cv-std : 0.0234640915273316
cv-max : 0.9174866722767857
cv-min : 0.8521969986100624
********** randomForestRegressor ************
Test score is 0.91
cross validatioion score
cv-mean : 0.8875629270273377
cv-std : 0.022124577367158674
cv-max : 0.9035613442825255
cv-min : 0.8453962100132185
********** gradientBoostRegressor ************
Test score is 0.91
cross validatioion score
cv-mean : 0.8839800036476907
cv-std : 0.02452673305433398
cv-max : 0.9140544186904889
cv-min : 0.8538996260972368
********** decisionTreeRegressor ************
Test score is 0.78
cross validatioion score
cv-mean : 0.8056426060069135
cv-std : 0.0222669686539792
cv-max : 0.8370426378836642
cv-min : 0.7780716563023613
********** knnRegressor ************
Test score is 0.66
cross validatioion score
cv-mean : 0.6436772448865766
cv-std : 0.03344162192431788
cv-max : 0.6893663835880288
cv-min : 0.6067558661871446
Top Performing models
1#print deceent models
2modelEvaluator.printDecentModels()
Top performing models are:
extraTreeRegressor score 0.924243991305789
randomForestRegressor score 0.9140009203234163
gradientBoostRegressor score 0.914941707121063
Hyper Parameter Tuning
Hyper parameters for top models
1for topModel in modelEvaluator.decentModels:
2 model = modelEvaluator.algorithms[topModel]
3 print("")
4 print("***************************")
5 print(" Model {} ".format(topModel))
6 print(model.get_params())
7 print("***************************")
8 print("")
9
10# We see all regressor has ~similar attributes hence we shall
11# use same cutomizing parameters for them
***************************
Model extraTreeRegressor
{'bootstrap': False, 'ccp_alpha': 0.0, 'criterion': 'mse', 'max_depth': None, 'max_features': 'auto', 'max_leaf_nodes': None, 'max_samples': None, 'min_impurity_decrease': 0.0, 'min_impurity_split': None, 'min_samples_leaf': 1, 'min_samples_split': 2, 'min_weight_fraction_leaf': 0.0, 'n_estimators': 100, 'n_jobs': None, 'oob_score': False, 'random_state': None, 'verbose': 0, 'warm_start': False}
***************************
***************************
Model randomForestRegressor
{'bootstrap': True, 'ccp_alpha': 0.0, 'criterion': 'mse', 'max_depth': None, 'max_features': 'auto', 'max_leaf_nodes': None, 'max_samples': None, 'min_impurity_decrease': 0.0, 'min_impurity_split': None, 'min_samples_leaf': 1, 'min_samples_split': 2, 'min_weight_fraction_leaf': 0.0, 'n_estimators': 100, 'n_jobs': None, 'oob_score': False, 'random_state': None, 'verbose': 0, 'warm_start': False}
***************************
***************************
Model gradientBoostRegressor
{'alpha': 0.9, 'ccp_alpha': 0.0, 'criterion': 'friedman_mse', 'init': None, 'learning_rate': 0.1, 'loss': 'ls', 'max_depth': 3, 'max_features': None, 'max_leaf_nodes': None, 'min_impurity_decrease': 0.0, 'min_impurity_split': None, 'min_samples_leaf': 1, 'min_samples_split': 2, 'min_weight_fraction_leaf': 0.0, 'n_estimators': 100, 'n_iter_no_change': None, 'presort': 'deprecated', 'random_state': None, 'subsample': 1.0, 'tol': 0.0001, 'validation_fraction': 0.1, 'verbose': 0, 'warm_start': False}
***************************
General overview
GridSearch technique is an exhaustive searching technique for hyperparameters hence it is reelatively slower.
RandomSearch searches for optimal hyperemeter randomly hence it is quite fast.
Parameter Tuning
1param_regressor = {
2 'bootstrap': [True, False],
3 'max_depth': [int(x) for x in np.linspace(5, 96, num = 2)],
4 'max_features': ['auto', 'log2'],
5 'min_samples_leaf': [1, 2, 4, 6, 8, 10, 12, 16, 32, 48],
6 'min_samples_split': [2, 4, 6, 8, 10, 12, 15],
7 'n_estimators': [int(x) for x in np.linspace(start = 2 , stop = 512, num = 2)]
8}
9
10param_gb_regressor = {
11 'max_depth': [int(x) for x in np.linspace(5, 96, num = 2)],
12 'max_features': ['auto', 'log2'],
13 'min_samples_leaf': [1, 2, 4, 6, 8, 10, 12, 16, 32, 48],
14 'min_samples_split': [2, 4, 6, 8, 10, 12, 15],
15 'n_estimators': [int(x) for x in np.linspace(start = 2 , stop = 512, num = 2)]
16}
RandomSearchCV
1modelEvaluator.runRandomSearchCV_HyperParameterTuningPipelines(param_regressor, param_gb_regressor)
Running randomsearch cv hyper parameter pipelines
********** extraTreeRegressor ************
Best parameter {'n_estimators': 512, 'min_samples_split': 2, 'min_samples_leaf': 6, 'max_features': 'auto', 'max_depth': 5, 'bootstrap': True}
Best score is 0.778791435146383
********** randomForestRegressor ************
Best parameter {'n_estimators': 512, 'min_samples_split': 4, 'min_samples_leaf': 8, 'max_features': 'log2', 'max_depth': 96, 'bootstrap': True}
Best score is 0.847613175720529
********** gradientBoostRegressor ************
Best parameter {'n_estimators': 512, 'min_samples_split': 8, 'min_samples_leaf': 4, 'max_features': 'log2', 'max_depth': 5}
Best score is 0.9511423708146253
GridSearchCV
1modelEvaluator.runGridSearchCV_HyperParameterTuningPipelines(param_regressor, param_gb_regressor)
Running gridsearch cv hyper parameter pipelines
********** extraTreeRegressor ************
Best parameter {'bootstrap': False, 'max_depth': 96, 'max_features': 'auto', 'min_samples_leaf': 1, 'min_samples_split': 4, 'n_estimators': 512}
Best score is 0.9242964126142897
********** randomForestRegressor ************
Best parameter {'bootstrap': False, 'max_depth': 96, 'max_features': 'log2', 'min_samples_leaf': 1, 'min_samples_split': 2, 'n_estimators': 512}
Best score is 0.9274128760044592
Conclusion
Initially, we applied available regression techniques which were available. Among them we found out the topmost model.
Using hyper paramter tuning we are able to get a boost in accuracy for from 0.2% to 0.4%
Data
Source Code
feature-engineering-solution.ipynb
comments powered by Disqus